Today, I'll talk about scientific stories from the book 'Bioinformatics Algorithms'.
This book may be beneficial for students like me.
This is because it can be an effective way to gain knowledge of bioinformatics and encourage algorithmic thinking.



To begin, I'll discuss Chapter 01 of 'Bioinformatice Algorithms' today.
If you find any errors or mistakes in the post, please send your comments to me via email.
E-mail address: tkm1214@naver.com
I'm looking forward to improving my posts with your advice.
1. The story of "DNA replication"
As you know, our body's blueprint is encoded in DNA.
DNA is the most important element in our body because it can be sent to the next generation as the blueprint.

If you spread it out, you will recognize that it consists of simple alphabets, such as A, C, G, and T.
Differences in people's genetic information are derived from the sequences of these bases.
And, you know, these sequences have contributed to human survival on Earth.

As the book mentioned, finding the ORI is important for understanding or utilizing the replication of DNA.
DNA information can be simplified using alphabets such as A, C, G, and T.
Let's talk about ways to find hidden messages located in DNA sequences.
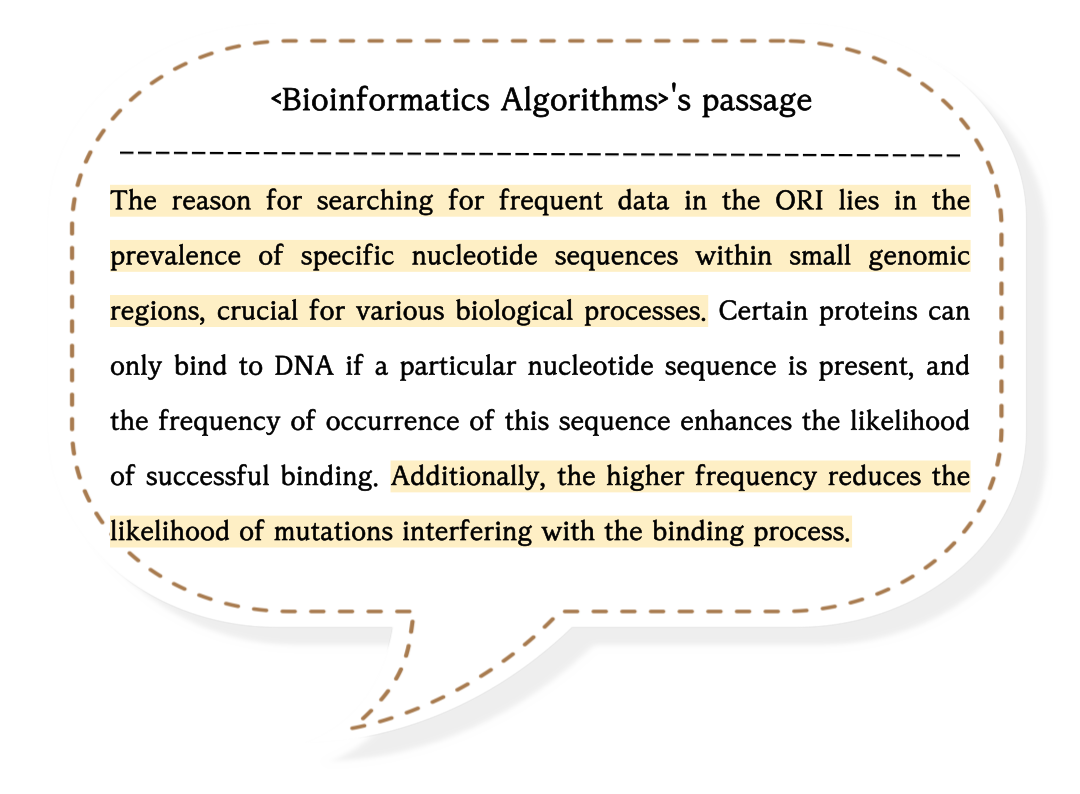
There are explanations for several algorithms to find frequent k-mer patterns in this book.
K-mer means 'String of length k'. So, if k is 4, ACCG can be a 4-mer pattern.
According to the book, protein 'DnaA' starts replication when it binds to a short sequence, 'DnaA box'.
So, in order to comprehend or utilize DNA replication effectively, it is essential to locate the DnaA box.
However, we may have to find the box without recognizing what the pattern is.
It's not easy, but if identical sequences are consistently found within a specific genomic region, it may suggest the presence of a potential DnaA box.

Generally, adenine (A) and guanine (G) form complementary pairs with thymine (T) and cytosine (C), respectively.
Then, have you heard about the reverse complementary pairs?
It is created by reversing the sequences of complementary bases.
For example, the complementary pair of 'ACG' is a 'TGC', and its reverse complement is a 'CGT'.
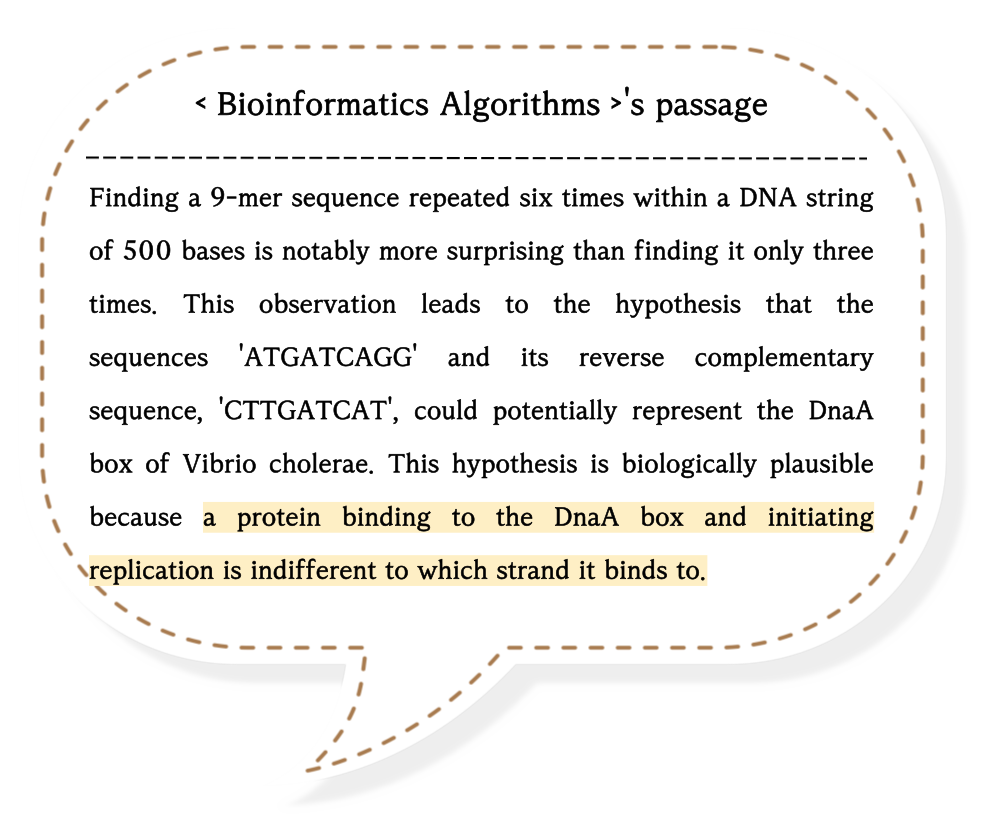
I spent a lot of time trying to understand the last sentence ↑, and I finally grasped it by drawing images.
Then, let's talk about the following image of DNA:

It's hard to understand the whole DNA replication process unless you're not majoring in biology.
So, I will introduce the process simply for you and for me.
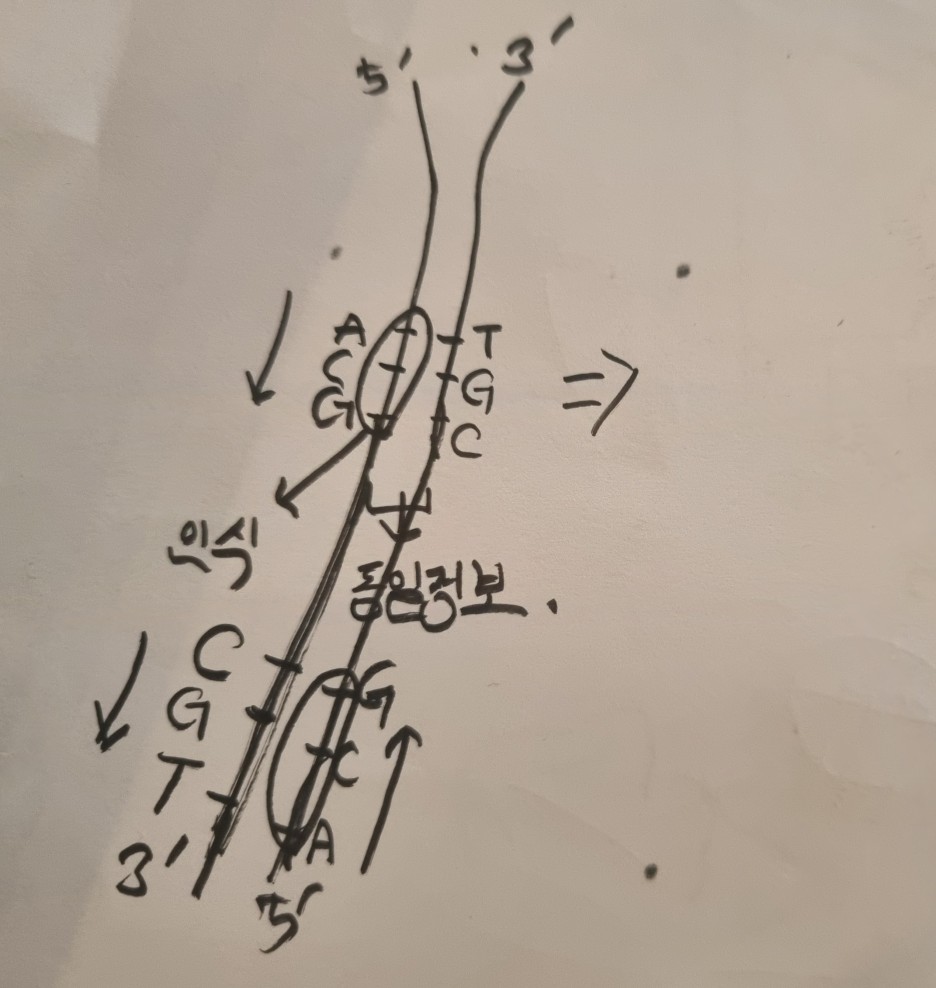
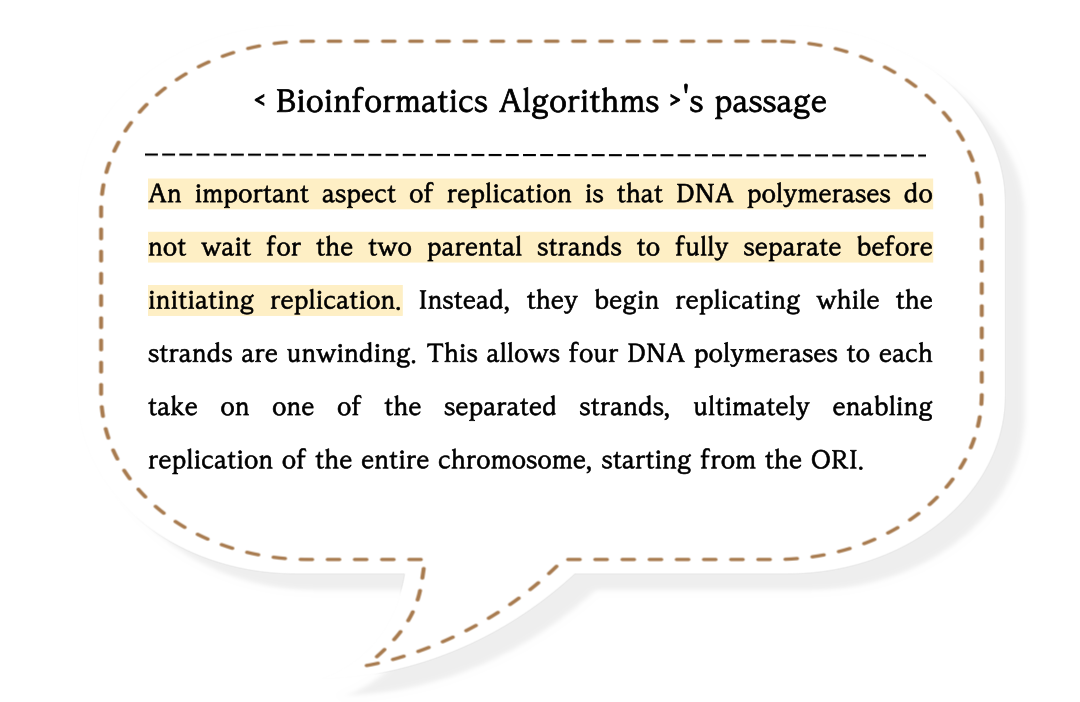
First of all, it's important to remember that DNA replication proceeds only in the 5' to 3' direction.
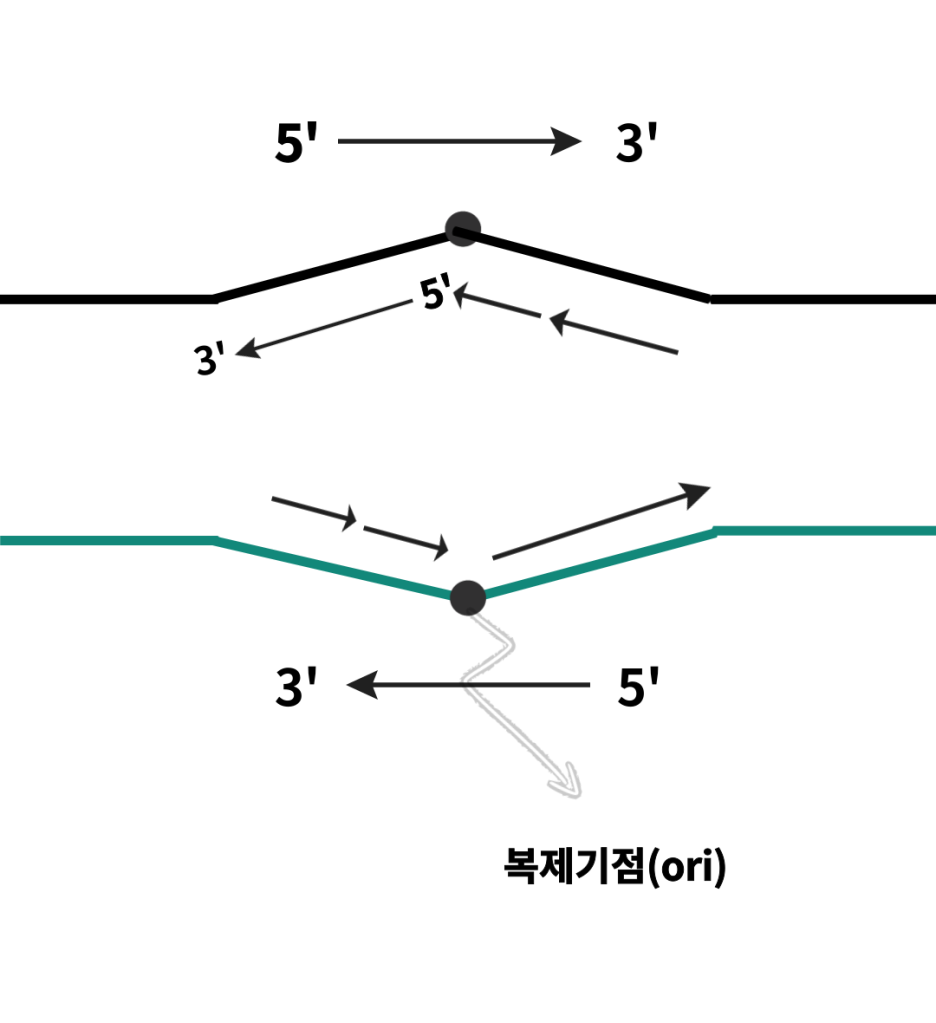
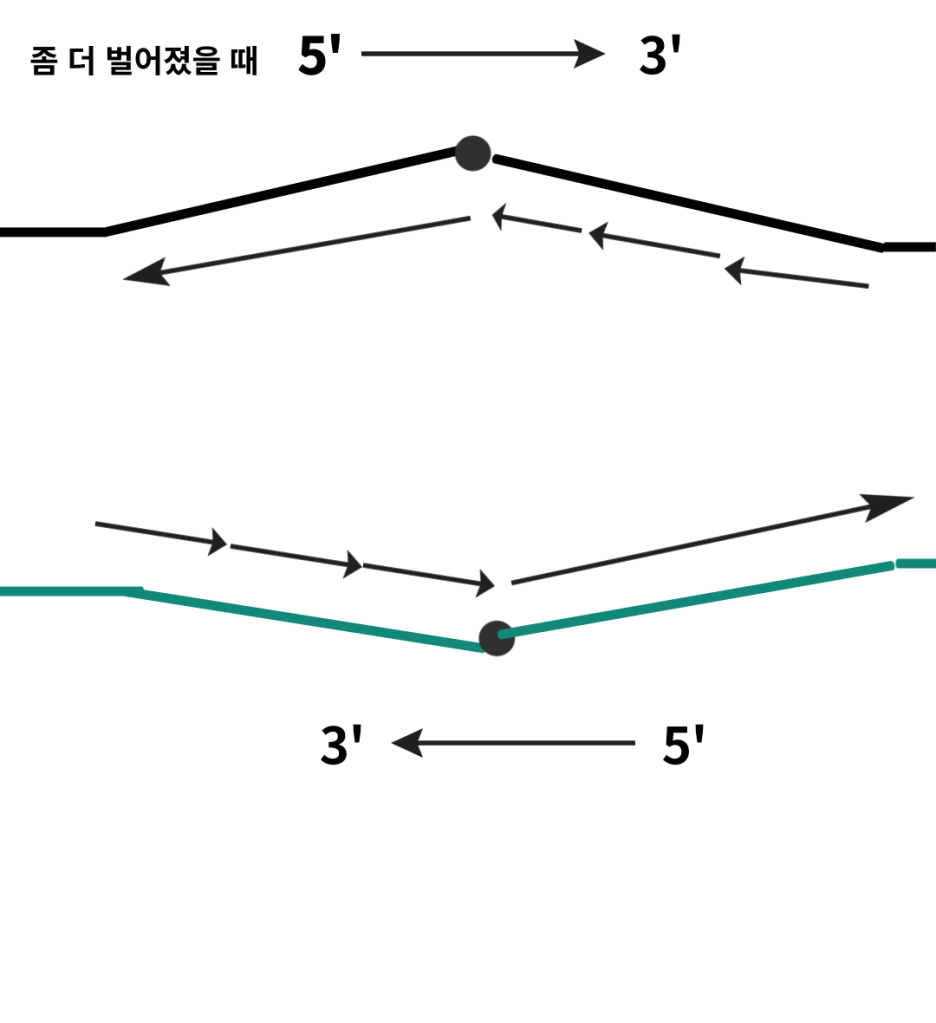
The presence of multiple arrows may cause confusion.
These are the results of a replication stop.
If you want to learn more, I recommend this video for you ↓
Replication stops can lead to some problems.
To be specific, a replication stop causes one half-strand to remain as a single strand for an extended period.
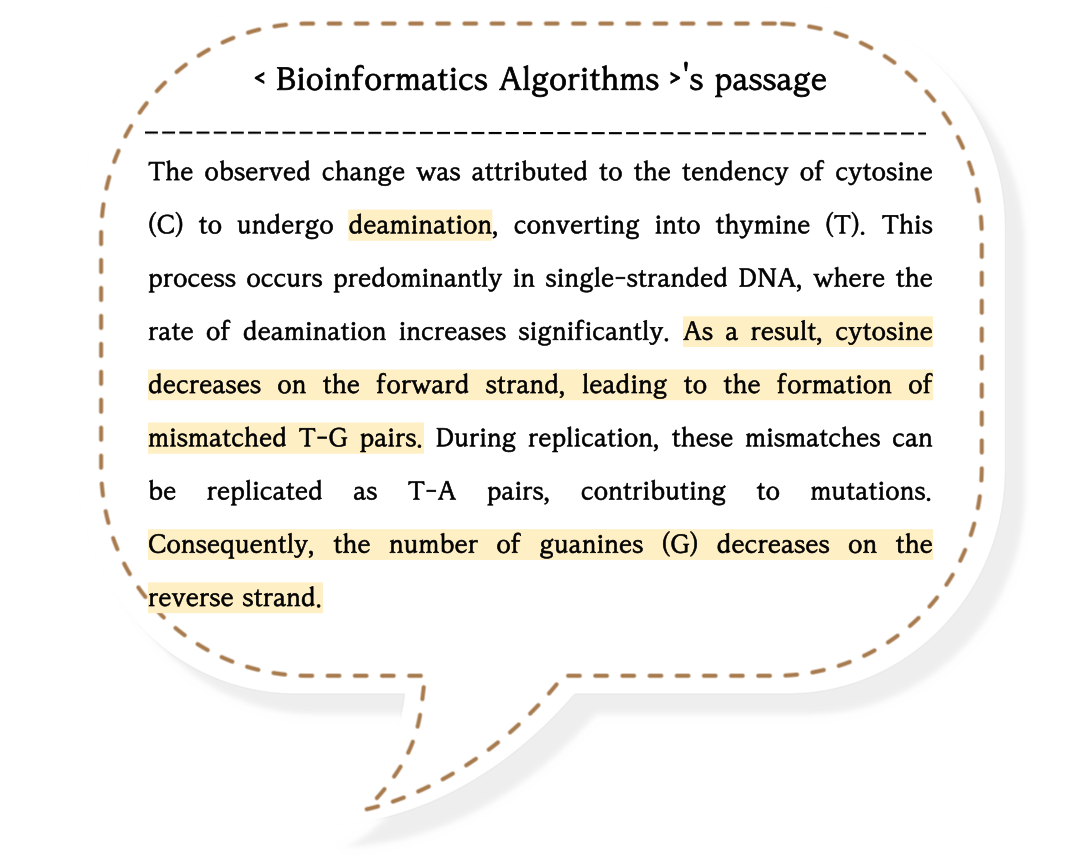
The difference in ratios between G and C can be used to locate the ORI.
Look at the image ↓
From ori to ter, 5' -> 3' is called a 'forward half-strand', and 3' -> 5' is called a reverse half-strand.
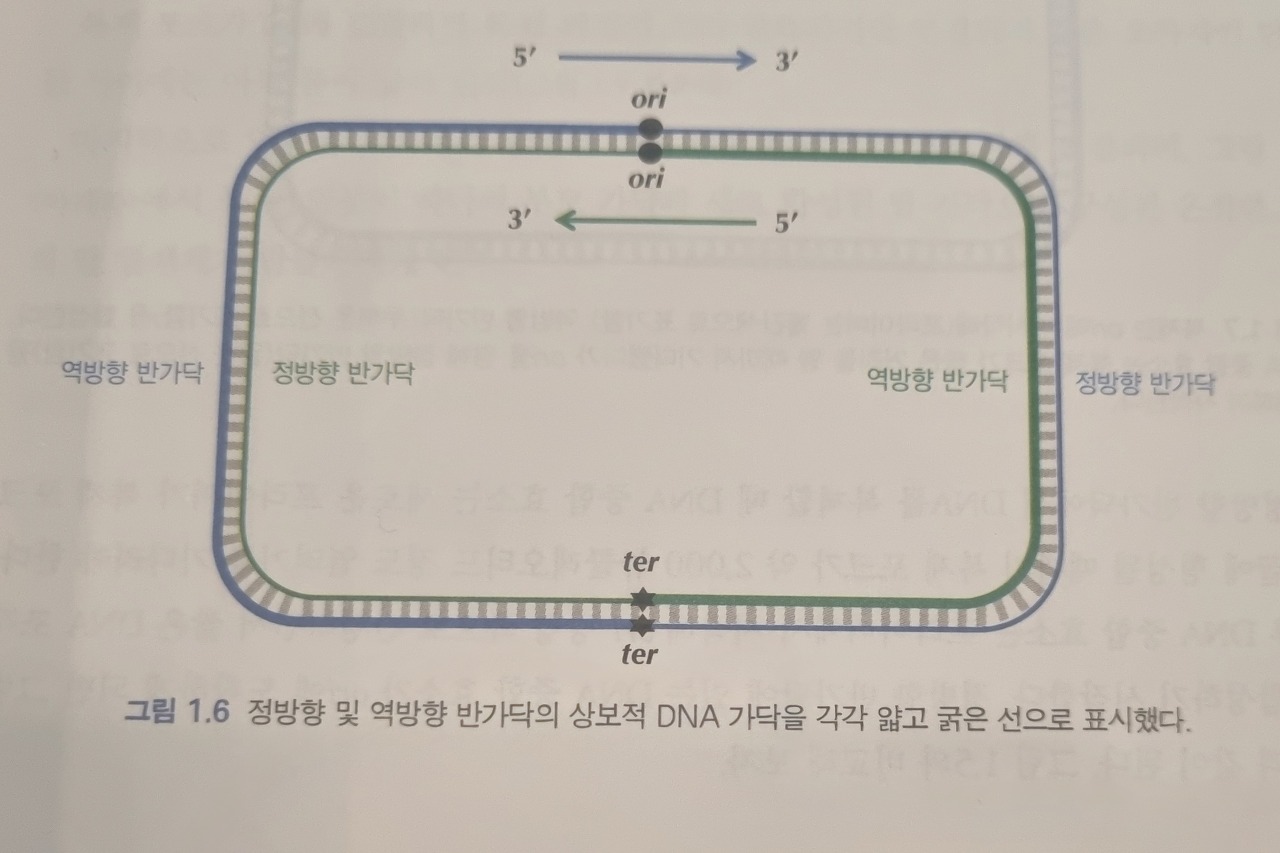
Due to deamination, the frequency of 'C' on the forward half-strand would decrease as a result of replication stops.
And that of 'G' on the reverse half-strand would decrease for the same reason.

By computing the difference in frequencies of nucleotides along the sequences, we can identify the location of the origin of replication (ORI).
Then, let's look at some algorithms mentioned in the book next time.
Bioinformatics OPEN STUDY | Notion
본 페이지는 비영리 목적으로 ‘생물정보학’ 관련 프로그래밍 공부 내용을 정리 및 공유하기 위해 마련해본 공간입니다. 본 페이지는 원래 비공개 페이지였지만, 일부를 공개 내용으로 바꾸는
sparkling-dibble-7ff.notion.site
과학 is 일상 : 네이버 블로그
과학기술정책가를 꿈꾸는 생물정보학도이자 과학기술계와 일반 대중을 연결하는 과학커뮤니케이터 TKM입니다! PC로 보면 좋습니다~ 공지 글로 과학꿈터뷰 전자책을 무료 공유하고 있습니다! 문
blog.naver.com




